Introduction
This page presents a performance evaluation for LDIF using life science use cases.
The following data sources have been used to evaluate the performance of LDIF:
- Allen Mouse Brain Atlas - a growing collection of online public resources integrating extensive gene expression and neuroanatomical data;
- KEGG GENES - a collection of gene catalogs for all complete genomes generated from publicly available resources;
- KEGG Pathway - a collection of pathway maps representing knowledge on the molecular interaction and reaction networks;
- PharmGKB - which provides data on gene information, disease and drug pathways, and SNP variants;
- Uniprot - a dataset containing protein sequence, genes and function.
Use case
In this use case, we evaluated the performance of LDIF integrating two larger life science datasets and translate those into a common target vocabulary.
Datasets
For this use case we used only KEGG GENES and UniProt datasets. There is a huge difference in dataset size between the two datasets. Converted to N-Triples the complete KEGG GENES dump is about 28GB in size whereas the UniProt dataset contains over 400GB worth of data.
For the test, we generated subsets of both data sources amounting together to 25, 100, 150 and 300 million RDF triples. The 3690 M dataset include the complete UniProt and complete KEGG GENES datasets.
Details about the benchmark datasets are summarized in the following table. It provides statistics about the data integration process for each dataset. The original number of input triples decreases in the process as LDIF discards input triples which are irrelevant for the defined mappings, and therefore can not be translated into the target vocabulary. The number decreases again after the actual translation, as the input data uses more verbose vocabularies and as multiple triples from the input data are thus combined into single triples in the target vocabulary. The size of the final dataset is the number of quads after the mapping phase plus any provided provenance data.
| 25M | 100M | 150M | 300M | 3690M | |
|---|---|---|---|---|---|
| Number of input quads | 25,000,000 | 100,000,000 | 150,000,000 | 300,000,000 | 3,687,918,681 |
| Number of quads after irrelevance filter | 13,576,394 | 44,249,757 | - | - | 1,164,250,713 |
| Number of quads after mapping | 4,419,410 | 24,972,112 | - | 32,211,677 | 98,380,001 |
| Number of pairs of equivalent entities resolved | 24,782 | 213,062 | - | 1,084,808 | 6,321,750 |
| Overall file size | 5.6 GB | 23 GB | 35 GB | 67 GB | - |
| Download link | 25M.zip | 100M.zip | - | - | - |
Mappings
We defined R2R mappings for translating genes, diseases and pathways from KEGG GENES and genes from UniProt into a proprietary target vocabulary. Some more sophisticated mappings from the use case translate complex structural patterns and perform value transformations (e.g. extracting an integer value from a URI). The prevalent value transformations are extracting strings with a regular expression and modifying the target data types.
Here are some examples of the mappings we used for:- mapping a KEGG GENE gene into a target vocabulary Gene
mp:Gene
a r2r:ClassMapping;
r2r:prefixDefinitions
"category: <http://mywiki/resource/category/> .
property: <http://mywiki/resource/property/> .
pathway: <http://wiking.vulcan.com/neurobase/kegg_pathway/resource/vocab/> .
genes: <http://wiking.vulcan.com/neurobase/kegg_genes/resource/vocab/> .
xsd: <http://www.w3.org/2001/XMLSchema#> .";
r2r:sourcePattern "?SUBJ a genes:gene";
r2r:targetPattern "?SUBJ a category:Gene";
. - mapping a KEGG GENE externalLink property into a target vocabulary UniprotId property
mp:GeneLinkUniProt
a r2r:PropertyMapping;
r2r:mappingRef mp:Gene;
r2r:sourcePattern "?SUBJ genes:externalLink ?x";
r2r:transformation "?id = regexToList('UniProt:(.+)', ?x)";
r2r:targetPattern "?SUBJ property:UniprotId ?'id'^^xsd:string";
.
Link Specifications
We defined the following Link Specification for the identity resolution phase:
<Silk>
<Prefixes>
<Prefix id="rdf" namespace="http://www.w3.org/1999/02/22-rdf-syntax-ns#" />
<Prefix id="rdfs" namespace="http://www.w3.org/2000/01/rdf-schema#" />
<Prefix id="owl" namespace="http://www.w3.org/2002/07/owl#" />
<Prefix id="property" namespace="http://mywiki/resource/property/" />
<Prefix id="category" namespace="http://mywiki/resource/category/" />
</Prefixes>
<Interlinks>
<Interlink id="link">
<LinkType>owl:sameAs</LinkType>
<SourceDataset dataSource="Source" var="a">
<RestrictTo>
{ ?a rdf:type category:Gene }
UNION { ?a rdf:type category:Disease }
UNION { ?a rdf:type category:Pathway }
</RestrictTo>
</SourceDataset>
<TargetDataset dataSource="Target" var="b">
<RestrictTo>
{ ?b rdf:type category:Gene }
UNION { ?b rdf:type category:Disease }
UNION { ?b rdf:type category:Pathway }
</RestrictTo>
</TargetDataset>
<LinkageRule>
<Aggregate type="max">
<Compare metric="equality">
<Input path="?a/property:UniprotId" />
<Input path="?b/property:UniprotId" />
</Compare>
<Compare metric="equality">
<Input path="?a/property:EntrezGeneId" />
<Input path="?b/property:EntrezGeneId" />
</Compare>
<Compare metric="equality">
<Input path="?a/property:MgiMarkerAccessionId" />
<Input path="?b/property:MgiMarkerAccessionId" />
</Compare>
<Compare metric="equality">
<Input path="?a/property:KeggDiseaseId" />
<Input path="?b/property:KeggDiseaseId" />
</Compare>
<Compare metric="equality">
<Input path="?a/property:KeggPathwayId" />
<Input path="?b/property:KeggPathwayId" />
</Compare>
</Aggregate>
</LinkageRule>
<Filter threshold="1.0" />
</Interlink>
</Interlinks>
</Silk>
Benchmark Machines
We used a machine with the following specification for the benchmark experiments:
FUB machines
These machines have been used for In-Memory, TDB and FUB Hadoop clusters.
| Hardware |
Processors: Intel i7 950, 3.07GHz (quadcore)
Memory: 24GB Hard Disks: 2 × 1.8TB (7,200 rpm) SATA2 |
| Software |
Operating System: Ubuntu 11.04 64-bit, Kernel: 2.6.38-10
Java version: 1.6.0_22 |
EC2 c1.medium instances
There machines have been used for EC2 Hadoop clusters.
| Hardware | Processor: 5 EC2 Compute Units1
Memory: 1.7 GB Hard Disks: 350 GB I/O Performance: Moderate |
| Software | Operating System: Ubuntu 11.04 32-bit |
Hadoop clusters
In all cluster configurations, the master works as job tracker and name node, while the slaves work as data nodes and task trackers.
FUB Hadoop 2-slaves cluster
- 1 master, 2 slaves (FUB machines)
- Network: Gigabit Ethernet
EC2 Hadoop X-slaves cluster
- 1 master, X slaves (EC2 c1.medium instances)
Test Procedure
We used LDIF version 0.5 both for the in-memory, TDB and Hadoop tests.
Single machine tests
For each single machine test, we applied the following procedure:
- Clear Operating System caches
echo 2 > /proc/sys/vm/drop_caches
- Run test
java -server -Xmx20G -jar ldif-single-machine.jar
Hadoop tests
For each distributed test, we applied the following procedure:
- Launch a master instance w/ EBS containing input data attached
- Launch slave instances
- Init the cluster (connect via SSH, clear all temporary data, format HDFS, run start-all)
- Upload input data into HDFS (from EBS)
- Run integration
Results
The following table summarizes the LDIF run times for the different dataset sizes. The overall run time is split according to the different processing steps of the integration process.
25M run times
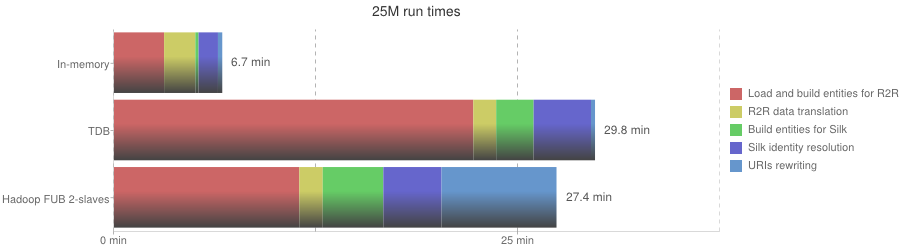
| Phase | In-memory | TDB | Hadoop FUB 2-slaves |
|---|---|---|---|
| Load and build entities for R2R | 186.3 s | 1334 s | 688 s |
| R2R data translation | 116.4 s | 85 s | 87 s |
| Build entities for Silk | 12.9 s | 139 s | 225 s |
| Silk identity resolution | 72.6 s | 213 s | 216 s |
| URIs rewriting | 16.2 s | 15 s | 427 s |
| Overall execution | 6.7 min | 29.7 min | 27.4 min |
100M run times
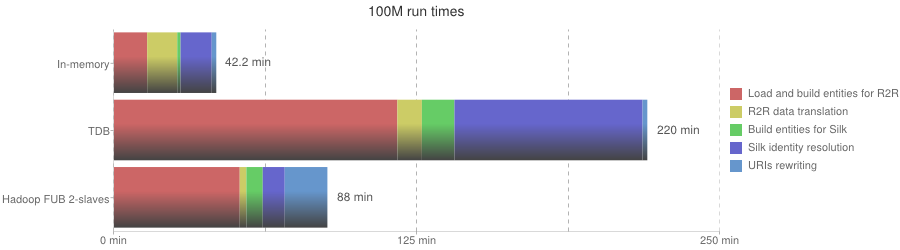
| Phase | In-memory | TDB | Hadoop FUB 2-slaves |
|---|---|---|---|
| Load and build entities for R2R | 822 s | 7014 s | 3112 s |
| R2R data translation | 744 s | 607 s | 165 s |
| Build entities for Silk | 81 s | 806 s | 405 s |
| Silk identity resolution | 772 s | 4656 s | 544 s |
| URIs rewriting | 113 s | 118 s | 1056 s |
| Overall execution | 42.2 min | 220 min | 88 min |
150M run times
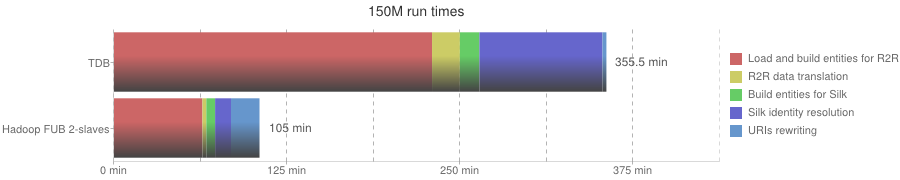
| Phase | In-memory | TDB | Hadoop FUB 2-slaves |
|---|---|---|---|
| Load and build entities for R2R | Out of Memory | 13776 s | 3830 s |
| R2R data translation | - | 1206 s | 170 s |
| Build entities for Silk | - | 847 s | 380 s |
| Silk identity resolution | - | 5328 s | 688 s |
| URIs rewriting | - | 173 s | 1235 s |
| Overall execution | - | 355 min | 105 min |
300M run times
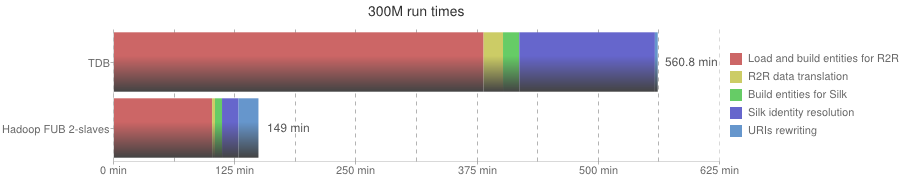
| Phase | In-memory | TDB | Hadoop FUB 2-slaves |
|---|---|---|---|
| Load and build entities for R2R | Out of Memory | 22870 s | 6070 s |
| R2R data translation | - | 1203 s | 179 s |
| Build entities for Silk | - | 1006 s | 436 s |
| Silk identity resolution | - | 8392 s | 1022 s |
| URIs rewriting | - | 176 s | 1232 s |
| Overall execution | - | 560 min | 148 min |
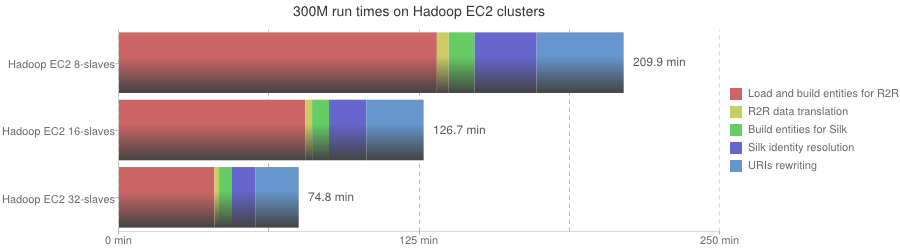
| Phase | Hadoop EC2 8-slaves | Hadoop EC2 16-slaves | Hadoop EC2 32-slaves |
|---|---|---|---|
| Load and build entities for R2R | 7933 s | 4647 s | 2382 s |
| R2R data translation | 297 s | 173 s | 114 s |
| Build entities for Silk | 646 s | 421 s | 324 s |
| Silk identity resolution | 1546 s | 932 s | 580 s |
| URIs rewriting | 2174 s | 1430 s | 1085 s |
| Overall execution | 209 min | 126 min | 75 min |
3690M run times
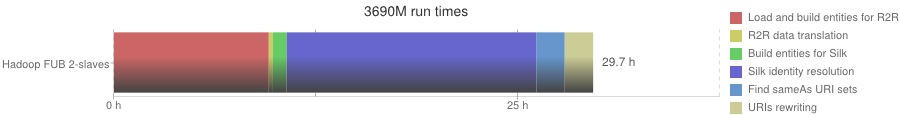
| Phase | Hadoop FUB 2-slaves (format: hh:mm:ss) |
|---|---|
| Load and build entities for R2R | 9:34:44 |
| R2R data translation | 15:22 |
| Build entities for Silk | 51:12 |
| Silk identity resolution | 15:27:52 |
| Find sameAs URI sets | 1:44:24 |
| URIs rewriting | 1:45:29 |
| Overall execution | 29:09:03 |
[1] According to this article, c1.medium instances with 5 ECU are observed to run as 2 of 4 cores of an Intel E5410 processor (4 cores, 2.33 GhZ).

